Project at the Korf Lab to find probable isoforms
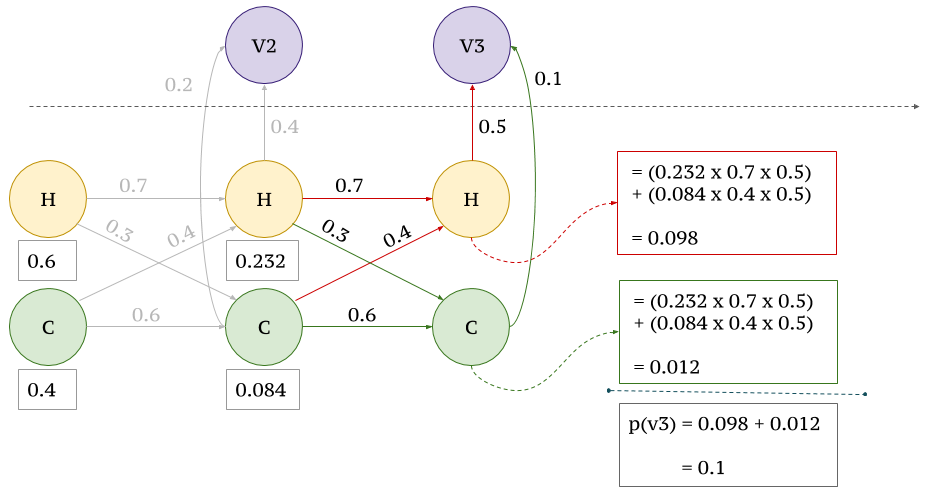
The same gene can be used to create multiple different gene products by splicing exons together in various ways through alternative splcing. These different products or transcripts are called isoforms. And of the possible transcripts, most are rare but due to the random and messy nature of splicing, rare isoforms do occur at a measurable rate. Isoforms that do not conform to our expectations of random splicing are more likely to have biological function and to find these isoforms, we used the Stochastic Viterbi algorithm. To find likely isoforms, our algorithm randomly traversed a Viterbi trellis, and the Forward-Backward algorithm was used to find the probability.
The goal of this algorithm is to find multiple, likely paths through a hidden Markov model. The algorithm is based on the Viterbi algorithm, but instead of finding the most likely path, it finds the n most likely paths. Probabilities of being in a state are calculated using a forward-backward algorithm.
The repo for this project can be found here.